About the Lab
Our goal is to understand the underlying principles of cellular responses to chromosomal insults and their roles in the maintenance of genomic stability.
Research Summary
Cancer is a complex disease driven by genetic and epigenetic alterations in the genome. To prevent these detrimental alterations, cells have evolved an intricate signaling network, called the DNA damage checkpoint, to detect and signal problems in the genome. During cancer development, the activation of oncogenes and loss of tumor suppressors leads to genomic instability, rendering cancer cells increasingly dependent upon specific DNA repair and checkpoint signaling proteins to survive. The Zou laboratory is particularly interested in understanding how the checkpoint detects DNA damage and genomic instability, and how the checkpoint can be targeted in cancer therapy. Our current studies are focused on understanding the molecular mechanisms by which different types of oncogenic events give rise to replication stress and genomic instability. Furthermore, we are developing new strategies to exploit the genomic instability and checkpoint addiction of different cancer cells in targeted cancer therapy.
Research Projects
ATM and ATR are two master checkpoint kinases in human cells. In particular, ATR is the key responder to a broad spectrum of DNA damage and DNA replication problems. We are especially interested in the mechanisms by which ATR is activated by replication stress and its functions in the replication stress response. Our recent studies have revealed that ATR plays an important role in the cellular responses to R loops, which arise from stable DNA:RNA hybrids during transcription. We found that ATR is activated by the collisions between replication forks and R loops, and it suppresses R loop-induced genomic instability through multiple mechanisms. We are extending our investigation to elucidate how ATR protects replication forks at R-loops and how ATR stabilizes the genome in response to aberrant R-loops.
The ATR checkpoint plays a key role in regulating and coordinating DNA replication, DNA repair, and cell cycle transitions. Recently, we have discovered a surprising function of ATR in mitosis. We have shown that ATR is localized to centromeres in mitosis, where it is activated by centromeric R loops. The activation of ATR at centromeres is critical for faithful chromosome segregation, thus revealing the unexpected importance of ATR in suppressing chromosomal instability (CIN). We have also developed new assays to understand how the alternative lengthening of telomere (ALT) pathway, which is regulated by ATR, is activated at telomeres. These new assays have helped us establish the framework of the ALT pathway for the first time, and uncovered the key mechanisms by which the ALT pathway is temporarily and spatially regulated during the cell cycle.
We are interested in the impacts of RNAs, including coding and non-coding RNAs, on genomic integrity. We recently discovered that RNA transcripts stimulate homologous recombination by forming a novel intermediate that contains both DNA:DNA and RNA:DNA hybrids. This intermediate, which we dubbed DR-loop, enhances the function of RAD51 in donor DNA. Our results demonstrate for the first time that RNA transcripts directly participate in DNA recombination, opening a new avenue to study the roles of RNA in DNA repair. These new findings will significantly change the current view of the functions of RNAs in DNA repair, providing new opportunities for cancer therapy.
During the evolution of tumors, cancer cells acquire mutations through a variety of mechanisms. We recently discovered that APOBEC3A/B proteins, two cytidine deaminases that are aberrantly expressed in multiple types of cancers, induce DNA replication stress and render cancer cells susceptible to ATR inhibition. Working with the team of Dr. Michael Lawrence, we find that APOBEC3A prefers substrate sites in DNA hairpins, leading to the discovery of passenger hotspot mutations in cancer. Furthermore, we find that the splicing factor mutations associated with myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML) induce R loops and trigger an ATR response. Cells that express these splicing factor mutants are sensitive to ATR inhibitors, providing a new strategy for the treatment of MDS and possibly other malignancies associated with RNA splicing defects.
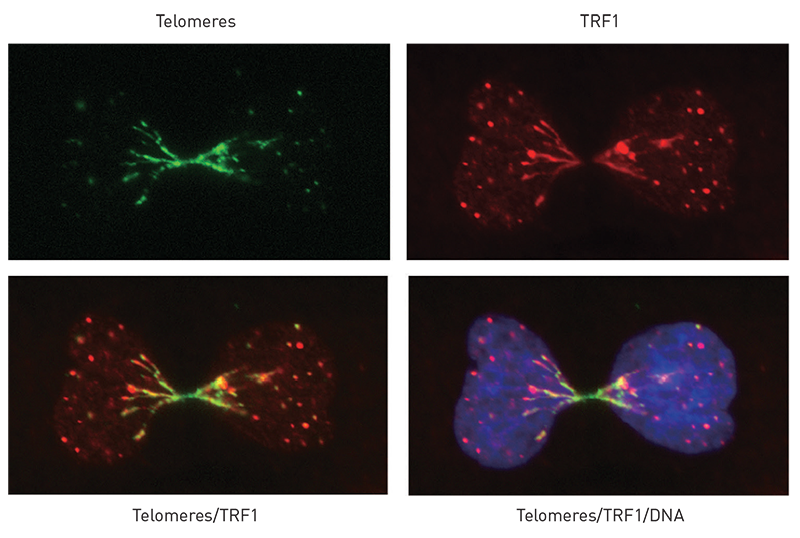
Research Image - Telomeric bridges in an ALT+ cancer cell lacking the BLM helicase.
Alternative lengthening of telomere (ALT) is a recombination-based mechanism to extend telomeres in cancer cells. We find that the BLM helicase is critical for resolving telomere recombination intermediates in ALT+ cancer cells. In the absence of BLM, unresolved recombination intermediates at telomeres result in chromosomal bridges in mitosis. Green: telomeres; Red: the telomere-binding protein TRF1; Blue: DNA.
Select Publications
Yadav T, Zhang J, Simoneau A, Ouyang J and Zou L. (2022) TERRA and RAD51AP1 promote alternative lengthening of Ttlomeres through an R-to-D loop switch. Mol. Cell. 82:3985-4000.
Yasuhara T, Xing Y-H, Bauer N, Lee L, Dong R, Yadav T, Soberman R, Rivera MN, and Zou L. (2022) Condensates induced by transcription inhibition localize active chromatin to nucleoli. Mol. Cell 82:2738-2753.
Genois MM, Gagné JP, Yasuhara T, Jackson J, Saxena S, Langelier MF, AhI, Bedford MT, Pascal JM, Vindigni A, Poirier GG, Zou L. CARM1 regulates replication fork speed and stress response by stimulating PARP1. Mol Cell. 2021 Feb 18;81(4):784-800.e8.
Zhang JM, Genois MM, Ouyang J, Lan L, Zou L. Alternative lengthening of telomeres is a self-perpetuating process in ALT-associated PML bodies. Mol Cell. 2021 Mar 4;81(5):1027-1042.e4.
Ouyang J, Yadav T, Zhang JM, Yang H, Rheinbay E, Guo H, Haber DA, Lan L, Zou L. RNA transcripts stimulate homologous recombination by forming DR-loops. Nature 2021 Jun;594(7862):283-288.
Simoneau A, Xiong R, Zou L. The trans cell cycle effects of PARP inhibitors underlie their selectivity toward BRCA1/2-deficient cells. Genes Dev. 2021 Sep 1;35(17-18):1271-1289.
Buisson R, Langenbucher A, Bowen D, Kwan EE, Benes CH, Zou L, and Lawrence SM. Passenger Hotspot Mutations in Cancer Driven by APOBEC3A and Mesoscale Genomic Features. Science 2019 Jun 28;364(6447):eaaw2872.
Kabeche, L., Nguyen, H. D., Buisson, R., and Zou, L. A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science 2018, 359:108-114.
Our Researchers
| Lab Member | Position | |
|---|---|---|
| Varsha Ananthapadmanabhan, Ph.D. | Postdoctoral Associate |  |
| Nolan Bick | Graduate Student (MCB) |  |
| Mariel De Los Santos | Graduate Student (Pharm) | |
| Morgan Feng | Undergraduate Student | |
| Xing Gao | Graduate Student (Pharm) | |
| Arijit Ghosh, Ph.D. | Postdoctoral Associate | |
| Yamin Gong, Ph.D. | Postdoctoral Associate |  |
| Ajinkya Kawale, Ph.D. | Research Associate, Senior |  |
| Xin Li, Ph.D. | Postdoctoral Associate | |
| Lin Lin, Ph.D. | Research Scientist | |
| Brenda Manzano-Winkler | Lab Research Analyst II |  |
| Yingying Meng, Ph.D. | Postdoctoral Associate | |
| Xiaojuan Ran, Ph.D. | Postdoctoral Associate | |
| Jiangchuan Shen, Ph.D. | Research Associate, Senior | |
| Yunhao Song, Ph.D. | Postdoctoral Associate |  |
| Yu Xu, Ph.D. | Research Scientist |
Lab Fun















